
 |
Processing multi-crystal datasets with xia2 and xia2.multiplex at SSRLThe program
Running
| ||||||||||||||||||||
pipeline=dials |
tell xia2 to use DIALS with DIALS scaling (default) |
pipeline=dials-aimless |
tell xia2 to use DIALS and AIMLESS scaling |
pipeline=3d |
tell xia2 to use XDS and XSCALE scaling, indexing with peaks found in a subset of images |
pipeline=3dii |
tell xia2 to use XDS and XSCALE scaling, indexing with peaks found from all images
|
Thus, the command line becomes something like:
xia2 --pipeline=3d /folder/with/image/files
This will start xia2, turn on the XDS pipeline, process all images found in the /folder/with/image/files path and then use XSCALE for scaling and merging.
In most cases this will be all you need, as xia2 will automatically do all the processing and return an acceptable result.
To evaluate the result, inspect the output in the terminal window, or the xia2.txt log file. A Table 1-like summary of the merging statistics is found at the end of the output or log file.
A more in-depth look at the processing results can be obtained by viewing a website-like summary of the run by issuing
firefox xia2.html
In cases where multiple imagesets (i.e. sets of images that are named using the same template, e.g. image_file_1_#####.cbf, where the hashes represent a number) are stored in the same folder, it is sufficient to point xia2 to that folder:
xia2 --pipeline=dials /folder/with/image/files
If these imagesets are scattered over several specific subfolders, the command can point to each imageset individually by specifying the first file in the set:
xia2 --pipeline=dials image=/folder/with/images/1/filename_1_00001.cbf image=/folder/with/images/2/filename_2_00001.cbf image=/folder/with/images/3/filename_3_00001.cbf
If selecting a subset of images is desired (for example, due to radiation damage), it’s possible to restrict processing to the specified image ranges:
xia2 --pipeline=dials image=/folder/with/images/1/filename_1_00001.cbf:1:40 /folder/with/images/2/filename_2_00001.cbf:1:35 /folder/with/images/3/filename_3_00001.cbf:1:45
Finally, one can feed a script to xia2, saved in a file with an .xinfo extension, which specifies all of the paths, files, and image ranges in one place. A typical .xinfo file looks like this:
BEGIN PROJECT AUTOMATIC BEGIN CRYSTAL DEFAULT BEGIN WAVELENGTH NATIVE WAVELENGTH 0.979500 END WAVELENGTH NATIVE BEGIN SWEEP SWEEP1 WAVELENGTH NATIVE DIRECTORY /folder/with/images/1/ IMAGE filename_1_0001.cbf START_END 1 40 END SWEEP SWEEP1 BEGIN SWEEP SWEEP2 WAVELENGTH NATIVE DIRECTORY /folder/with/images/2/ IMAGE filename_2_0001.cbf START_END 1 35 END SWEEP SWEEP2 BEGIN SWEEP SWEEP3 WAVELENGTH NATIVE DIRECTORY /folder/with/images/3/ IMAGE filename_3_0001.cbf START_END 1 45 END SWEEP SWEEP3 END CRYSTAL DEFAULT END PROJECT AUTOMATIC
The .xinfo file is automatically generated the first time xia2 is run; conversely, the above template can be used to create one from scratch. The file can then be submitted to xia2 and it will read all of its information from :
xia2 --info=info.xinfo
For best results a multi-crystal dataset should be processed iteratively, in several cycles, with each cycle featuring adjustments that improve the merging statistics. While the procedure varies for each dataset, below is a general approach that we found to be useful.
This run serves two purposes: to weed out the non-processable imagesets (or "sweeps") and to generate the *.xinfo file that can be used for subsequent runs. For the purposes of this manual, we are going to assume that all images are located in the same folder.
xia2 --failover=True --read_all_image_headers=False --multi_crystal=True /folder/with/image/files multiprocessing.mode=parallel multiprocessing.njob=4 multiprocessing.nproc=4
The options prior to the path to image files (designated with double dashes) are used to set xia2-specific parameters; here, the options are selected to speed up data processing, as multi-crystal sets can take a while.
failover=True |
skip sweeps that cause errors without terminating the entire run |
read_all_image_headers=False |
assume that one image header applies to the entire sweep, this saves time |
multi_crystal=True |
tell xia2 that you are using data from multiple crystals |
After the path to files, you can add more options to the command line; note that these are not designated with the double-dash. The multiprocessing options will cause xia2 to run in parallel. NOTE: running xia2 in parallel ensures that errors to processing one sweep do not terminate the whole run.
multiprocessing.mode=parallel |
activate parallel processing mode |
multiprocessing.njob=4 |
number (e.g 4) of sweeps to be processed in parallel |
multiprocessing.nproc=4 |
number (e.g. 4) of CPUs to use per sweep |
The automatic.xinfo file is generated upon the first launch of xia2 (from step 1); it is advisable to not overwrite this version, but create a working copy:
cp automatic.xinfo work.xinfo
The file can then be edited as desired using the text editor of preference. (An app called geany is implemented on all pxproc machines and features a user-friendly GUI.)
The most common edits to the *.xinfo file include changing the image ranges within a sweep or deleting a problematic sweep altogether; after editing and saving the file, xia2 can be rerun as follows:
xia2 --info=work.xinfo
It is useful to not only delete the sweep on which xia2 had failed, but also note which sweeps were not successfully indexed, as well. A telltale sign of such a failure could be found in the xia2 output (on screen and in the xia2.txt log file) and would look like this:
------------------- Autoindexing SWEEP8 --------------------
All possible indexing solutions:
tP 58.64 58.64 151.42 90.00 90.00 90.00
oC 82.86 82.92 151.40 90.00 90.00 90.00
oP 58.61 58.65 151.39 90.00 90.00 90.00
mC 82.69 82.84 151.21 90.00 89.91 90.00
mP 58.62 58.60 151.33 90.00 89.99 90.00
aP 58.52 58.56 151.21 89.95 89.95 90.07
Indexing solution:
tP 58.64 58.64 151.42 90.00 90.00 90.00
-------------------- Integrating SWEEP8 --------------------
Processed batches 3 to 22
Standard Deviation in pixel range: 0.47 0.55
Integration status per image:
oooooooo............
"o" => good "%" => ok "!" => bad rmsd
"O" => overloaded "#" => many bad "." => weak
"@" => abandoned
Mosaic spread: 0.060 < 0.060 < 0.060
-------------------- Spotfinding SWEEP9 --------------------
184 spots found on 20 images (max 17 / bin)
**
**
** *
** **
**** *** *
**** *** * * ***
******** **** ******
********************
********************
********************
2 image 21
------------------- Autoindexing SWEEP9 --------------------
Processing sweep SWEEP9 failed: No suitable indexing solution could be found.
You can view the reciprocal space with:
dials.reciprocal_lattice_viewer /folder/to/processing/DEFAULT/NATIVE/SWEEP9/index/108_optimised.expt /folder/to/processing/DEFAULT/NATIVE/SWEEP9/index/106_SWEEP9_strong.refl
In the above fragment of the log, notice that Sweep 8 indexed and integrated normally; conversely, Sweep 9 failed to index, with an error message. Based on this information, one could edit work.xinfo to remove Sweep 9 from further processing.
One could also try and see how the processing would work with more information about the crystal parameters extracted from this initial attempt.
Inspecting the output, one could see that quite a few of the sweeps (if not all of them) are indexed in the same symmetry, with highly similar unit cell parameters, e.g.:
------------------- Autoindexing SWEEP8 -------------------- All possible indexing solutions: tP 58.64 58.64 151.42 90.00 90.00 90.00 oC 82.86 82.92 151.40 90.00 90.00 90.00 oP 58.61 58.65 151.39 90.00 90.00 90.00 mC 82.69 82.84 151.21 90.00 89.91 90.00 mP 58.62 58.60 151.33 90.00 89.99 90.00 aP 58.52 58.56 151.21 89.95 89.95 90.07 Indexing solution: tP 58.64 58.64 151.42 90.00 90.00 90.00
It is possible to re-run xia2 with these specific crystal parameters, which will aid the processing by stabilizing the indexing of datasets and improving the chances of success.
xia2 --info=work.xinfo --failover=True --read_all_image_headers=False space_group=P4 unit_cell=58.64,58.64,151.42,90.00,90.00,90.00
(note the lack of dashes before space_group and unit_cell options!)
By this point, some iteration of steps 2-3 should produce a completed xia2 run, which will present the summary output on screen and write it out to the xia2.txt log file. Parts of this output can be inspected to see if any sweeps should be omitted, usually because they decrease the quality of the merging statistics for the entire dataset.
One useful piece of information is the estimated resolutions for each sweep, which can be found towards the end of the processing run, and look like this:
--------------------- Scaling DEFAULT ----------------------
Resolution for sweep NATIVE/SWEEP7: 1.78 (cc_half > 0.3)
Resolution for sweep NATIVE/SWEEP15: 1.76 (cc_half > 0.3)
* Resolution for sweep NATIVE/SWEEP11: 2.39 (cc_half > 0.3)
Resolution for sweep NATIVE/SWEEP3: 1.77 (cc_half > 0.3)
Resolution for sweep NATIVE/SWEEP5: 1.59 (cc_half > 0.3)
Resolution for sweep NATIVE/SWEEP4: 1.53 (cc_half > 0.3)
Resolution for sweep NATIVE/SWEEP2: 1.71 (cc_half > 0.3)
* Resolution for sweep NATIVE/SWEEP9: 3.91 (cc_half > 0.3)
Resolution for sweep NATIVE/SWEEP14: 1.64 (cc_half > 0.3)
Resolution for sweep NATIVE/SWEEP10: 1.96 (cc_half > 0.3)
Resolution for sweep NATIVE/SWEEP1: 1.79 (cc_half > 0.3)
Resolution for sweep NATIVE/SWEEP6: 1.63 (cc_half > 0.3)
Resolution for sweep NATIVE/SWEEP13: 1.69 (cc_half > 0.3)
Resolution for sweep NATIVE/SWEEP12: 1.50 (cc_half > 0.3)
* Resolution for sweep NATIVE/SWEEP8: 0.00
Note that for most of the sweeps, the resolution varied between ~1.5Å and ~2Å, but three datasets (which we marked with asterisks) seem to be the outliers. Two of them have resolution of 2.4Å and 3.9Å, while the third one seems to be of such poor quality that its resolution is not even estimated.
xia2 will also create a xia2.html file, which can be used to view the results as charts. The most useful - for this type of data - chart can be found under the "Dataset NATIVE" tab. (The "NATIVE" name is the default for your dataset, you can change it in the work.xinfo file to something else.)
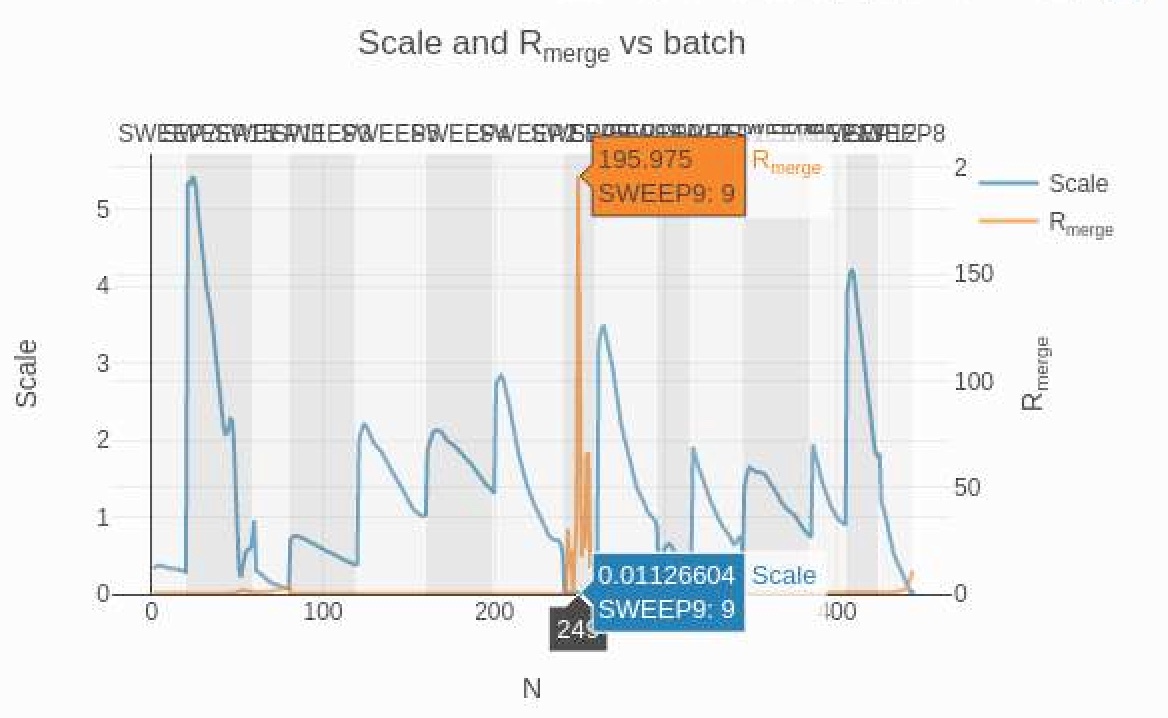
This tab will show the Table 1 summary (same as the one under the "Summary" tab), the Xtriage warnings (if any), and below them, Analysis Plots. Clicking on "Analysis by batch" will expose several useful plots that will show if any sweeps act as obvious outliers. In the example dataset, the graph of Rmerge and scale vs. frame shows a concerningly high Rmerge for Sweep 9. (If sweep names overlap too much, mousing over any point of the chart will reveal which sweep the concerning Rmerge belongs to.)
The high multiplicity of this dataset allows us to edit the work.xinfo file to exclude Sweep 9 and re-run processing without it. The next run may reveal that more sweeps have problematic statistics and they, too can be removed from processing by editing the work.xinfo file. We can also remove sweeps with resolutions that are too low for our purposes, such as Sweeps 11 and 8, judging by the resolution chart above. We can continue this process until we are satisfied with the merging statistics.
xia2.multiplexxia2.multiplex [2] is a program that improves a multi-crystal dataset by performing symmetry analysis, re-scaling and re-merging, as well as analyzing the various pathologies that bedevil multi-crystal datasets, such as non-isomorphism, radiation damage, and cases of preferential crystal orientation (which results in missing cones of data). This program can be run on the entirety of the merged dataset from above, or on selected sweeps if that is desired. xia2.multiplex accepts a set of two types of files: the reflection table file (with the extension .refl) and a file with stored experiment metadata (with the extension .expt). These files (both for each sweep and for the entire dataset) can be found in the DataFiles subfolder of your xia2 processing directory. Thus, to run xia2.multiplex on the data you just processed, issue
xia2.multiplex DataFiles/AUTOMATIC_DEFAULT_scaled.expt DataFiles/AUTOMATIC_DEFAULT_scaled.refl
One could also select several sweeps to see if a better complete dataset can be assembled from those, e.g.:
xia2.multiplex DataFiles/AUTOMATIC_DEFAULT_NATIVE_SWEEP1.expt DataFiles/AUTOMATIC_DEFAULT_NATIVE_SWEEP1.refl DataFiles/AUTOMATIC_DEFAULT_NATIVE_SWEEP3.expt DataFiles/AUTOMATIC_DEFAULT_NATIVE_SWEEP3.refl DataFiles/AUTOMATIC_DEFAULT_NATIVE_SWEEP7.expt DataFiles/AUTOMATIC_DEFAULT_NATIVE_SWEEP7.refl
xia2.multiplex will also perform a Laue check of the point group for this crystal and ensure that the dataset is isomorphous. If some sweeps are found to be outliers (i.e. if multiple crystal forms are present), this information could be used to separate the datasets.
It is more useful to monitor the multiplicity-independent Rpim rather than the multiplicity-dependent Rmerge or Rmeas, since the latter can be cosmetically manipulated by reducing multiplicity, i.e. throwing data away. However, in cases of multi-crystal datasets, it is sometimes useful to sacrifice some multiplicity in exchange for gains in overall resolution or for tangible statistical improvements. In cases of crystals that have high-symmetry space groups, it is very easy to amass a highly-redundant dataset and thus to have the option to pick out only the best (in terms of resolution, mosaicity, etc.) sweeps, rather than include everything.
xia2.multiplex performs hierarchical clustering of datasets to determine which sweeps best match up with one another. This approach is very useful in cases of non-isomorphism, as it very effectively separates different isoforms.
Cos(angle) clustering summary: ========= ============== =================== ======== ============== ============== Cluster No. datasets Datasets Height Multiplicity Completeness ========= ============== =================== ======== ============== ============== 1 2 4 5 7.7e-09 5 0.93 2 2 7 9 6.8e-05 1.7 0.83 3 3 3 7 9 0.00022 4.3 1 4 2 1 8 0.00067 4.4 0.99 5 3 2 4 5 0.00081 7.7 0.98 6 4 0 3 7 9 0.0014 7.2 1 7 5 1 2 4 5 8 0.006 11.9 1 8 6 1 2 4 5 6 8 0.014 14.8 1 9 10 0 1 2 3 4 5 6 7 8 9 0.023 22 1 ========= ============== =================== ======== ============== ==============
Clustering can also be used to identify smaller subsets that could have better merging statistics than the full set of sweeps. In this particular case, a combination of datasets #0, 3, 7 and 9 may be useful to try. As you can see, the jump in the "Height" parameter from cluster 6 to cluster 7 is rather abrupt, the overall multiplicity is still a respectable 7.2, and the dataset composed of those sweeps would be 100% complete.
Unfortunately, the numbering scheme of the datasets does not directly correlate with the numbering scheme of the sweeps. To correlate one with the other, we need to open xia2.multiplex.html file in a web browser, expand the "Datasets" item, and look at which image filenames the datasets numbers correlate to: Here, dataset 0 correlates with files named "thaulmatin_1_#####.cbf", dataset 3 with "thaulmatin_13_#####.cbf", dataset 7 with "thaulmatin_15_#####.cbf", and dataset 9 with "thaulmatin_6_#####.cbf", or with Sweeps 4, 6, 7, and 12 respectively.
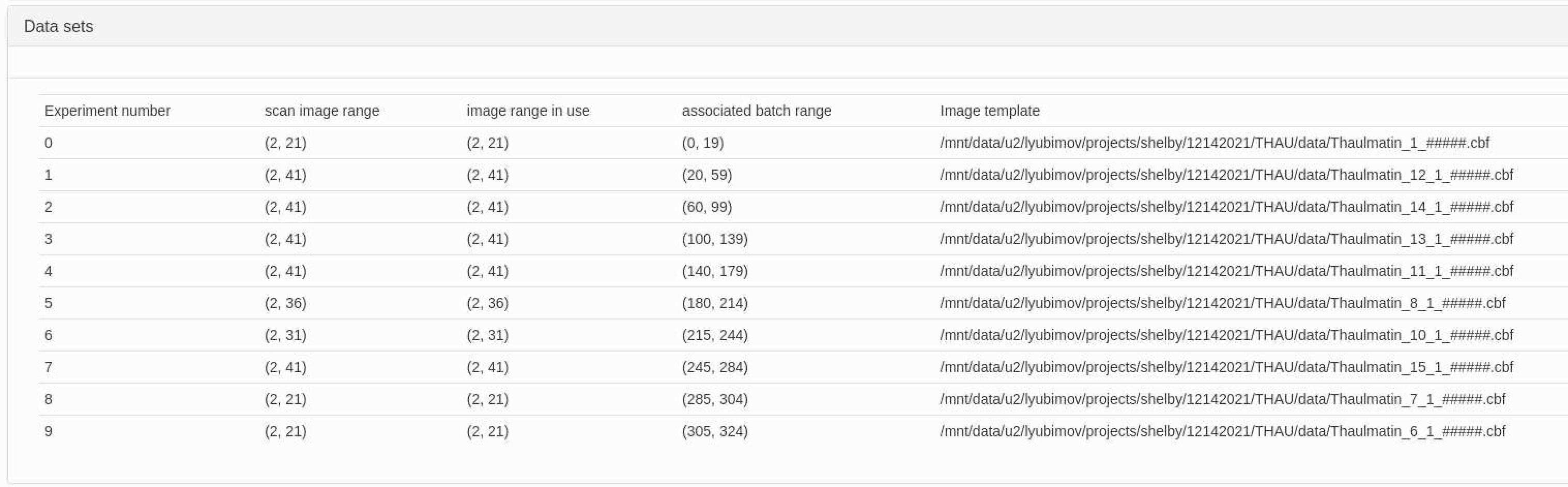
Once the corresponding sweeps are identified, the work.xinfo file can be edited to restrict the processing to these sweeps alone.
NOTE: In all steps "[username]" is a placeholder for your username. Make sure you replace all instances of it with your own username, otherwise nothing will work.
Step 1. Preparation
pxproc machine that appears the least used at the moment; we recommend pxproc1-12, as they contain more CPUs than the others.cd /data/[username]/
xia2 tutorial subfolder:
mkdir xia2_tutorial
cd xia2_tutorial
Step 2. Initial xia2 run.
xia2 at the sample HEWL data accessible via a symlink, and run it in multiprocessing mode as above:
xia2 --failover=False --read_all_image_headers=False --multi_crystal=True /data/[username]/templates/xia2_sample_data/ multiprocessing.mode=parallel multiprocessing.njob=10 multiprocessing.nproc=4
Step 3. Removing unhelpful sweeps
xia2.txt file; note that
Processing sweep SWEEP10 failed: xia2.integrate subprocess failed with exitcode 1) reported for Sweep 10.xia2.html using Firefox; click on the "Dataset NATIVE" tab and expand the "Analysis by batch" section. Note that
automatic.xinfo file:
cp automatic.xinfo work.xinfo
work.xinfo file to remove Sweep9 and Sweep 10 from processing. (We will deal with Sweeps 1 and 8 later.)xia2 using the exited work.xinfo:
xia2 --failover=False --read_all_image_headers=False --multi_crystal=True --info=work.xinfo multiprocessing.mode=parallel multiprocessing.njob=10 multiprocessing.nproc=4
Step 4. Removing unhelpful images
xia2.txt again and note that spotfinding results for all sweeps show some deterioration with exposure, but especially for Sweeps 1 and 8.xia2.html with Firefox, and inspect the "Analysis by batch" charts; note that
work.xinfo to truncate image ranges as follows:
Note: these are semi-arbitrary ranges; you can make your own decisions here.
xia2 with work.xinfo:
xia2 --failover=False --read_all_image_headers=False --multi_crystal=True --info=work.xinfo multiprocessing.mode=parallel multiprocessing.njob=10 multiprocessing.nproc=4
Step 5. Refining the merged dataset with xia2.multiplex
xia2.multiplex with the results of this latest processing run:
xia2.multiplex DataFiles/AUTOMATIC_DEFAULT_scaled.expt DataFiles/AUTOMATIC_DEFAULT_scaled.refl
xia2.multiplex are
expt extension containing information about the experimentrefl extension containing the reflection table xia2.multiplex.log file and find the "overall merging statistics". Note the improvement vs. the result of the previous xia2 processing run.xia2.multiplex.html file can be inspected in Firefox to see more helpful statistics and charts.